In recent weeks, I found myself going to the doctors to be treated for a largely benign but quite unappealing infection. Arriving there, they let me know almost immediately that it was impetigo, a common bacterial infection. Impetigo can be caused by two different bacterial species, Staphylococcus aureus (S. aureus) or Streptococcus pyogenes (S. pyogenes), both being Gram-positive bacteria. Thus, I was given a prescription for the steroidal antibiotic fusidic acid to rub in the affected area. This particular antibiotic works by binding to and inactivating an important bacterial protein, called EF-G-GDP. EF-G-GDP is known as an elongation factor, which functions in protein production. Elongation factors work in the ribosome where proteins are produced, helping to translate bacterial mRNA code into polypeptide chains (proteins). So, when fusidic acid binds to EF-G-GDP, this elongation factor cannot help translate mRNA code into proteins, which stops the bacteria from being able to replicate. Fusidic acid also prevents EF-G-GDP from disassembling the ribosome in the bacteria, another hindrance to bacterial protein production and therefore bacterial survival1. Antibiotic treatments like these are often vital for serious bacterial infections to be cleared, as well as for use during medical procedures like surgical operations to prevent infection of a patient. Even down to some of the meat products we eat, antibiotics are used to ensure animals are free of disease before being consumed by us. This makes antibiotics one of the most useful and important discoveries of the last century when it comes to our health.
With regards to the antibiotic treatment of my impetigo, there was a problem – the impetigo was getting worse. It could have been for many reasons: that I was spreading the infection around my skin by rubbing in the fusidic acid antibiotic cream, that the sores were constantly moist from the cream and being given an ideal environment for bacterial replication, or (most likely) how erratic my schedule of cream application was. Another trip to the doctor later and a second prescription was given, this time for flucloxacillin, a beta-lactam antibiotic that inhibits bacterial cell wall synthesis2, a process essential for bacterial survival. Within the week-long oral course, my impetigo was gone.
In retrospect, there may also be a case for antibiotic resistance contributing to the lack of efficacy shown by the fusidic acid antibiotic cream. Antibiotic resistance can occur in bacteria intrinsically (as it evolved to combat natural antibiotics secreted by other bacteria and fungi), through natural selection of genetic mutations that confer resistance to specific antibiotics3,4. A high genetic mutation rate (a driver of evolution, and therefore of developing antibiotic resistance mechanisms) in S. aureus has been seen when fusidic acid is the sole treatment for impetigo5, with mutation of the fusA gene (encoding EF-G-GDP, the aforementioned elongation factor), and the fusB and fusC genes (encoding cytoplasmic proteins that protect the target site of fusidic acid) being causes for this1. Luckily, impetigo isn’t a devastating infection when treatment is hindered by antibiotic resistance, asides from its annoyance. Though, this still made me think: what about antibiotic resistance in other, deadlier, diseases?
Resistance to antibiotics in human use is not a new concept. In fact, it’s been cropping up since the 1940s, when penicillin, the first antibiotic discovered in 1928, was being used as a treatment for bacterial infections6,7. A huge proportion of S. aureus infections had become penicillin-resistant by the late 1960s (more than 80%!). Even when semisynthetic methicillin had been introduced strains of S. aureus soon became resistant, becoming the methicillin-resistant S. aureus (MRSA) you might have heard of today8. This phenomenon looks like it’s only going to rise with time, with a predicted 10 million extra deaths due to antimicrobial resistance (including antibiotic resistance) per year by 2050 if global antibiotics consumption stays on its current course9, and 1.27 million deaths already directly attributed to bacterial antibiotic resistance being recorded in 201910. Antibiotic agents and antibiotic resistance genes have been shown to spread throughout the environment via wastewater from urban areas and via faeces from agricultural animals11,12, with no current limitations on the concentration of antibiotics entering the environment from wastewater in the UK13. Even non-resistant bacteria can obtain antibiotic resistance genes from other resistant bacteria, in a process called horizontal gene transfer (HGT)14. HGT can occur via conjugation, where bacterial plasmids are transferred between individual bacteria, transformation, where free DNA encoding antibiotic resistance from the environment is taken up by bacteria, or via lysogenic conversion. Lysogenic conversion occurs when DNA from a bacteriophage (a virus that infects bacteria) is inserted into the bacterial genome during infection, and is then repackaged back into the bacteriophage accidentally taking along with it an antibiotic resistance gene, to infect another bacterium. This way, antibiotic resistance spreads throughout the environment (Fig.1).
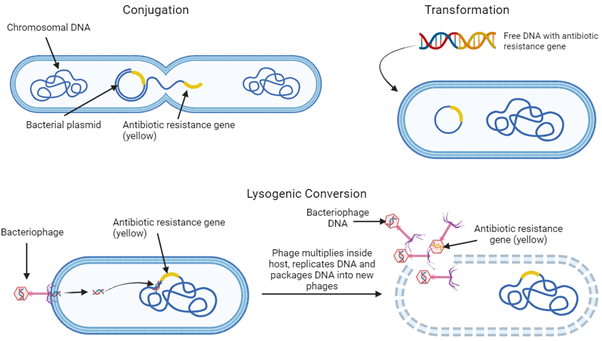
Figure 1: Giving to the needy – Horizontal Gene Transfer, the way genes are spread in their local bacterial environment.
A tempting solution to this seemingly catastrophic problem is to just pump more money into looking for more antibiotics that bacteria aren’t resistant to yet, and this would make sense initially. However, new antibiotic development has slowed in recent years15, and the endless march of evolution is likely to result in the same problems as before. Perhaps, then, the best option is to find a treatment that can evolve alongside bacteria. In this case, the way forwards takes us to a lesser-known solution from the past.
In 1917, the microbiologist Félix d’Herelle described “an invisible microbe” that could kill bacteria with much greater efficacy than any other methods known at the time. This was the bacteriophage, discovered by William Twort two years earlier (where Twort had not realized their antibacterial potential as d’Herelle did). Different bacteriophages, or phages, were seen to target specific bacterial species and strains, highlighting their vast medical potential16. This was a leap towards controlling bacterial infections that would previously have been fatal, and there was much international success in so-called “Phage therapy”, both disease-preventing and disease-curing, in the following 1920s and 1930s. The dawn of the age of antibiotics in the 1940s, however, soon relegated this innovative use of bacteria’s natural enemy to the past – at least in the West. To this day, phage therapy is being developed and used in Eastern Europe (with plausibility in the idea that Soviet development of phage therapy “tainted” the field, and dissuaded Western scientists from further contribution17). Is it time for researchers across the world to return to our neglected friend, the bacteriophage?
There seem to be a lot of benefits to this, at least with lytic phages (as these replicate in the bacteria until the host cell bursts or “lyses”, whereas lysogenic phages incorporate their DNA into the host genome and allow the bacteria to pass on the DNA to future generations, potentially spreading antibiotic resistance genes as mentioned before). Phages can eliminate bacterial infections without affecting human cells, and their levels increase over the course of treatment while antibiotic levels only decrease over the course of treatment. Additionally, they can be produced very easily at low cost, and there are many different phages that are specific to strains of bacterial species18,19. Phage specificity is a huge advantage to this therapy, as it means there is less need for broad-spectrum antibiotic use which can kill other, non-harmful bacteria in the gut and disrupt the gut microbiome. As phages still have the capacity to evolve, and co-evolve with their bacterial host20, there may be some changes to their efficacy as treatment. This, however, can be countered by the use of phage “cocktails”, where several different phages all specific to the same bacterial species are administered21. Furthermore, phage “training” is a novel concept also used to ensure phages can beat the bacteria. This involves leaving bacteria and phage together for extended periods of time, allowing them to co-evolve in a biological “arms race” that increases phage infectivity and host ranges (how many strains of a bacterial species they can infect), and reduces bacterial resistance of the original “ancestral” bacterial generation to this trained generation of phage22. This method used by Borin et al23 found promising results in their initial report in training phages against E. coli, and there’s potential for its use in developments of phage therapies.
For administration of phage therapy, there are many options. Historically administration of phage went through any of the following routes: topical, intramuscular, subcutaneous or intraperitoneal. But, in recent times researchers have found encapsulating phages in materials like alginate (a polymer derived from brown seaweed) to be an effective way to prepare phages to survive gastrointestinal conditions, like high acidity, for oral delivery into the gut24, meaning many gut-based infections could potentially be ameliorated. Even more interestingly, one team of researchers genetically engineered an E. coli-specific phage to express an E. coli outer membrane protein, allowing it to survive the gastrointestinal tract’s extreme acidity25. Further “designer phage” work includes modification of phage receptor binding proteins (proteins that allow phages to attach to bacteria), with this modification increasing phage host ranges26. Some animal trials of phage therapy have already happened with some success, including phage use against V. cholerae (the bacteria that causes cholera) in rabbits27. Clinical trials involving humans are ongoing, primarily in France and the United States, and mainly focus on E. coli, S. aureus, K. pneumonia, and P. aeruginosa28. Before phage-only therapies are much more widely available, success has been found in combining antibiotic and phage therapies against antibiotic-resistant bacteria. Bacteria maintaining resistance to both phage and antibiotics can often be costly to its resources, and so this combination therapy results in bacterial susceptibility to either treatment, a phenomenon known as Phage-Antibiotic Synergy (PAS)29.
There are current limitations to how far phage therapy can be adopted while still adhering to modern manufacturing standards, known as Good Manufacturing Practice (GMP)30. As phages are biological products capable of evolution, and phage treatments (including cocktails and trained phages) may be tailored to resolve specific bacterial infections of a single patient, following these high GMP standards for constantly updated bacteriophage treatments may be extremely time-consuming and costly31. Also, natural bacteriophages can be difficult to patent unless they are obviously genetically modified or are used in combination with other engineered phages31,32, as natural phages are not a novel invention. This may pose a problem for investment from pharmaceutical companies, as a lack of patentability of a treatment can disincentivize investment into its manufacturing. These concerns are yet to be resolved, although flexible medical licensing with regards to phage therapies would be helpful in getting specialized or personalized phage treatments into use quicker.
Recent developments in bacteriophage therapy all suggest seemingly limitless potential for its wider use in tackling our growing problem of antibiotic resistance. With more and more attention being focused on the once overlooked phage solution, we may be able to see a new “golden era” for phage therapies, similar to that celebrated by antibiotics in the previous decade, and overcome a pressing issue threatening modern medicine as we know it.
References
- Fernandes P. Fusidic Acid: A Bacterial Elongation Factor Inhibitor for the Oral Treatment of Acute and Chronic Staphylococcal Infections. Cold Spring Harb Perspect Med. 2016 Jan 4;6(1):a025437.
- Flucloxacillin [Internet]. [cited 2024 Feb 29]. Available from: https://pubchem.ncbi.nlm.nih.gov/compound/Flucloxacillin
- Clardy J, Fischbach MA, Currie CR. The natural history of antibiotics. Curr Biol. 2009 Jun 9;19(11):R437–41.
- Normark BH, Normark S. Evolution and spread of antibiotic resistance. J Intern Med. 2002 Aug;252(2):91–106.
- Brown EM, Thomas P. Fusidic acid resistance in Staphylococcus aureus isolates. Lancet. 2002 Mar 2;359(9308):803.
- Fleming A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. 1929. Bull World Health Organ. 2001;79(8):780–90.
- Crofton J, Mitchison DA. Streptomycin resistance in pulmonary tuberculosis. Br Med J. 1948 Dec 11;2(4588):1009–15.
- Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest. 2003 May;111(9):1265–73.
- O’Neill J. Tackling drug-resistant infections globally: final report and recommendations. Government of the United Kingdom [Internet]. 2016 May 19 [cited 2024 Feb 29]; Available from: https://apo.org.au/node/63983
- Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Aguilar GR, Gray A, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022 Feb 12;399(10325):629–55.
- Berglund B. Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics. Infect Ecol Epidemiol. 2015 Sep 8;5:28564.
- He Y, Yuan Q, Mathieu J, Stadler L, Senehi N, Sun R, et al. Antibiotic resistance genes from livestock waste: occurrence, dissemination, and treatment. npj Clean Water. 2020 Feb 19;3(1):1–11.
- Hayes A, May Murray L, Catherine Stanton I, Zhang L, Snape J, Hugo Gaze W, et al. Predicting selection for antimicrobial resistance in UK wastewater and aquatic environments: Ciprofloxacin poses a significant risk. Environ Int. 2022 Nov;169:107488.
- von Wintersdorff CJH, Penders J, van Niekerk JM, Mills ND, Majumder S, van Alphen LB, et al. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front Microbiol. 2016 Feb 19;7:173.
- Butler MS, Henderson IR, Capon RJ, Blaskovich MAT. Antibiotics in the clinical pipeline as of December 2022. J Antibiot. 2023 Aug;76(8):431–73.
- Fruciano DE, Bourne S. Phage as an antimicrobial agent: d’Herelle’s heretical theories and their role in the decline of phage prophylaxis in the West. Can J Infect Dis Med Microbiol. 2007 Jan;18(1):19–26.
- Summers WC. Bacteriophage Therapy. Annu Rev Microbiol. 2001 Oct 1;55(1):437–51.
- Carlton RM. Archivum Immunologiae et Therapiae Experimentalis. vol. 1999;47:267–74.
- Loc-Carrillo C, Abedon ST. Pros and cons of phage therapy. Bacteriophage. 2011 Mar;1(2):111–4.
- Koskella B, Brockhurst MA. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 2014 Sep;38(5):916–31.
- Gu J, Liu X, Li Y, Han W, Lei L, Yang Y, et al. A method for generation phage cocktail with great therapeutic potential. PLoS One. 2012 Mar 1;7(3):e31698.
- Betts A, Vasse M, Kaltz O, Hochberg ME. Back to the future: evolving bacteriophages to increase their effectiveness against the pathogen Pseudomonas aeruginosa PAO1. Evol Appl. 2013 Nov;6(7):1054–63.
- Borin JM, Avrani S, Barrick JE, Petrie KL, Meyer JR. Coevolutionary phage training leads to greater bacterial suppression and delays the evolution of phage resistance. Proc Natl Acad Sci U S A [Internet]. 2021 Jun 8;118(23). Available from: http://dx.doi.org/10.1073/pnas.2104592118
- Durr HA, Leipzig ND. Advancements in bacteriophage therapies and delivery for bacterial infection. Mater Adv. 2023 Mar 6;4(5):1249–57.
- Nobrega FL, Costa AR, Santos JF, Siliakus MF, van Lent JWM, Kengen SWM, et al. Genetically manipulated phages with improved pH resistance for oral administration in veterinary medicine. Sci Rep. 2016 Dec 15;6:39235.
- Lin TY, Lo YH, Tseng PW, Chang SF, Lin YT, Chen TS. A T3 and T7 recombinant phage acquires efficient adsorption and a broader host range. PLoS One. 2012 Feb 9;7(2):e30954.
- Bhandare S, Colom J, Baig A, Ritchie JM, Bukhari H, Shah MA, et al. Reviving Phage Therapy for the Treatment of Cholera. J Infect Dis. 2019 Feb 15;219(5):786–94.
- Hitchcock NM, Devequi Gomes Nunes D, Shiach J, Valeria Saraiva Hodel K, Dantas Viana Barbosa J, Alencar Pereira Rodrigues L, et al. Current Clinical Landscape and Global Potential of Bacteriophage Therapy. Viruses [Internet]. 2023 Apr 21;15(4). Available from: http://dx.doi.org/10.3390/v15041020
- Diallo K, Dublanchet A. Benefits of Combined Phage-Antibiotic Therapy for the Control of Antibiotic-Resistant Bacteria: A Literature Review. Antibiotics (Basel) [Internet]. 2022 Jun 22;11(7). Available from: http://dx.doi.org/10.3390/antibiotics11070839
- Bretaudeau L, Tremblais K, Aubrit F, Meichenin M, Arnaud I. Good Manufacturing Practice (GMP) Compliance for Phage Therapy Medicinal Products. Front Microbiol. 2020 Jun 4;11:1161.
- The antimicrobial potential of bacteriophages [Internet]. [cited 2024 Feb 29]. Available from: https://publications.parliament.uk/pa/cm5804/cmselect/cmsctech/328/report.html
- 2022 [cited 2024 Feb 29]. Patenting viruses and phages. Available from: https://www.dehns.com/patenting-viruses-and-phages/